iTRAQ in Quantitative Proteomics: Principles, Workflow, and Applications
-
Multi-sample study design and grouping
-
Optimization of reporter ion interference
-
Deep bioinformatics analysis
-
Integrative multi-omics workflows
In comparative proteomic studies involving multi-sample analyses, disease mechanism investigations, and drug target discovery, iTRAQ-based quantitative proteomics has gained widespread adoption due to its high throughput capability, analytical sensitivity, and experimental reproducibility.
Principles Underlying iTRAQ-Based Quantitative Proteomics: Achieving Multi-Sample Protein Quantification
iTRAQ is an isobaric tagging strategy for peptide-based relative quantification in mass spectrometry. Peptides originating from different biological samples are derivatized using distinct isotopic tags that share identical masses at the MS1 level. Upon MS/MS fragmentation, these tags generate reporter ions of different m/z values, enabling relative quantification based on reporter ion intensities.
1. Chemical Architecture of iTRAQ Reagents
Each iTRAQ reagent consists of three functional components:
(1) Reporter group (114-121 Da), used to distinguish sample channels.
(2) Balance group, maintaining equivalent total mass across tags.
(3) Peptide-reactive group, targeting peptide N-termini and lysine side chains.
The isobaric design ensures that all labeled peptides remain mass-identical in MS1, whereas reporter ions generated in MS2 allow downstream quantitative discrimination.
2. Experimental Workflow for iTRAQ-Based Proteomics
(1) Protein extraction followed by trypsin digestion.
(2) Differential labeling of peptide samples using iTRAQ tags (e.g., 114, 115, 116…).
(3) Mixing of labeled samples and LC fractionation coupled to high-resolution LC-MS/MS analysis.
(4) Generation of reporter ions during MS/MS fragmentation and quantification via reporter ion signal intensities.
(5) Data processing involving peptide identification, protein annotation, and differential protein expression analyses.
Applications of iTRAQ-Based Quantitative Proteomics
1. Mechanistic Studies of Disease Biology
Comparative profiling of protein expression between healthy and disease conditions allows for the identification of differentially expressed proteins, supporting biomarker discovery and pathway-level mechanistic investigation.
2. Drug Mechanism-of-Action and Target Identification
Protein expression changes between drug-treated and control groups provide insights into drug target engagement and downstream signaling pathways, informing preclinical drug development.
3. Developmental and Temporal Profiling Studies
iTRAQ enables labeling of samples collected across developmental stages or experimental time points, allowing dynamic tracking of protein expression changes in contexts such as embryogenesis or cellular differentiation.
4. Integrative Multi-Omics Analyses
iTRAQ is frequently integrated with metabolomics, transcriptomics, and related omics platforms to construct multi-layered molecular networks and identify central regulatory modules in complex biological systems.
MtoZ Biolabs additionally supports integrative analyses combining iTRAQ with phosphoproteomics, metabolomics, and immunoproteomics datasets to facilitate systems-level modeling.
Technical Limitations of iTRAQ and Corresponding Mitigation Strategies
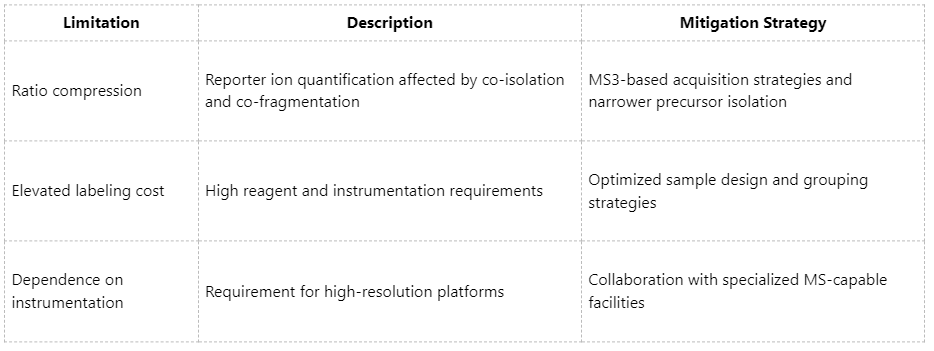
As a mature multi-channel quantitative proteomics platform, iTRAQ continues to play a significant role in life sciences research, pharmaceutical investigations, and translational studies. Its high-throughput, parallel sample quantification capabilities are well suited for differential protein expression analyses, molecular network reconstruction, and biomarker validation.
MtoZ Biolabs provides comprehensive iTRAQ-based quantitative proteomics services spanning sample preparation, iTRAQ labeling, high-resolution LC–MS/MS acquisition, and downstream data analysis with visualization. Tailored strategies are further offered for:
Researchers are welcome to contact MtoZ Biolabs for support in advancing ongoing projects efficiently.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







